Usage and installation of shinyChromosome
This is the repository for the Shiny application presented in “shinyChromosome: An R/Shiny Application for Interactive Creation of Non-circular Plots of Whole Genomes” (Yu et al. Genomics Proteomics Bioinformatics. 2020).
Use shinyChromosome online
shinyChromosome is deployed at http://150.109.59.144:3838/shinyChromosome/, http://shinychromosome.ncpgr.cn/ and https://yimingyu.shinyapps.io/shinychromosome/ for online use.
shinyChromosome is idle until you activate it by accessing the URLs.
So, it may take some time when you access this URL for the first time.
Once it was activated, shinyChromosome could be used smoothly and easily.
Launch shinyChromosome directly from R and GitHub
User can choose to run shinyChromosome installed on local computers (Windows, Mac or Linux) for a more preferable experience.
Step 1: Install R and RStudio
Before running the app you will need to have R and RStudio installed (tested with R 3.5.0 and RStudio 1.1.419).
Please check CRAN (https://cran.r-project.org/) for the installation of R.
Please check https://www.rstudio.com/ for the installation of RStudio.
Step 2: Install the R Shiny package and other packages required by shinyChromosome
Start an R session using RStudio and run these lines:
# try an http CRAN mirror if https CRAN mirror doesn't work
install.packages("shiny")
install.packages("rlang")
install.packages("zip")
install.packages("ggplot2")
install.packages("plyr")
install.packages("ggthemes")
install.packages("RLumShiny")
install.packages("RColorBrewer")
install.packages("gridExtra")
install.packages("reshape2")
install.packages("data.table")
install.packages("shinythemes")
install.packages("shinyBS")
install.packages("markdown")
# install shinysky
install.packages("devtools")
devtools::install_github("venyao/ShinySky", force=TRUE)
Step 3: Start the app
Start an R session using RStudio and run these lines:
shiny::runGitHub("shinyChromosome", "venyao")
This command will download the code of shinyChromosome from GitHub to a temporary directory of your computer and then launch the shinyChromosome app in the web browser. Once the web browser was closed, the downloaded code of shinyChromosome would be deleted from your computer. Next time when you run this command in RStudio, it will download the source code of shinyChromosome from GitHub to a temporary directory again. This process is frustrating since it takes some time to download the code of shinyChromosome from GitHub.
Users are suggested to download the source code of shinyChromosome from GitHub to a fixed directory of your computer, such as ‘E:\apps’ on Windows. Following the procedure illustrated in the following figure, a zip file named ‘shinyChromosome-master.zip’ would be downloaded to the disk of your computer. Move this file to ‘E:\apps’ and unzip this file. Then a directory named ‘shinyChromosome-master’ would be generated in ‘E:\apps’. The scripts ‘server.R’ and ‘ui.R’ could be found in ‘E:\apps\shinyChromosome-master’.
Then you can start the shinyChromosome app by running these lines in RStudio.
library(shiny)
runApp("E:/apps/shinyChromosome-master", launch.browser = TRUE)
Deploy shinyChromosome on local or web Linux server
Step 1: Install R
Please check CRAN (https://cran.r-project.org/) for the installation of R.
Step 2: Install the R Shiny package and other packages required by shinyChromosome
Start an R session and run these lines in R:
# try an http CRAN mirror if https CRAN mirror doesn't work
install.packages("shiny")
install.packages("rlang")
install.packages("zip")
install.packages("ggplot2")
install.packages("plyr")
install.packages("ggthemes")
install.packages("RLumShiny")
install.packages("RColorBrewer")
install.packages("gridExtra")
install.packages("reshape2")
install.packages("data.table")
install.packages("shinythemes")
install.packages("shinyBS")
install.packages("markdown")
# install shinysky
install.packages("devtools")
devtools::install_github("venyao/ShinySky", force=TRUE)
For more information, please check the following pages:
https://cran.r-project.org/web/packages/shiny/index.html
https://github.com/rstudio/shiny
https://shiny.rstudio.com/
Step 3: Install Shiny-Server
Please check the following pages for the installation of shiny-server.
https://www.rstudio.com/products/shiny/download-server/
https://github.com/rstudio/shiny-server/wiki/Building-Shiny-Server-from-Source
Step 4: Upload files of shinyChromosome
Put the directory containing the code and data of shinyChromosome to /srv/shiny-server.
Step 5: Configure shiny server (/etc/shiny-server/shiny-server.conf)
# Define the user to spawn R Shiny processes
run_as shiny;
# Define a top-level server which will listen on a port
server {
# Use port 3838
listen 3838;
# Define the location available at the base URL
location /shinychromosome {
# Directory containing the code and data of shinyChromosome
app_dir /srv/shiny-server/shinyChromosome;
# Directory to store the log files
log_dir /var/log/shiny-server;
}
}
Step 6: Change the owner of the shinyChromosome directory
$ chown -R shiny /srv/shiny-server/shinyChromosome
Step 7: Start Shiny-Server
$ start shiny-server
Now, the shinyChromosome app is available at http://IPAddressOfTheServer:3838/shinyChromosome/.
Input data format
The detailed format of input data for different types of plots are described in the following sections.
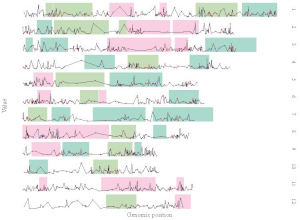
1. Single-genome plot
1.1 Genome data
The dataset should contain only 2 columns with fixed order. Column names are optional.
1st column: chromosome ID.
2nd column: chromosome length.
Acceptable input data format can be
chr size
1 43268879
2 35930381
3 36406689
or
1 43268879
2 35930381
3 36406689
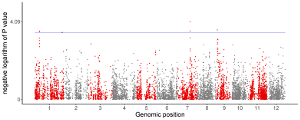
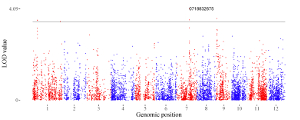
1.2 Point
The dataset should contain >=3 columns.
In the simplest situation, the dataset should contain 3 columns with fixed order. In this case, column names are optional.
1st column: chromosome ID.
2nd column: chromosome position.
3rd column: data value.
Acceptable input data format can be
chr position value
1 202360 0.315323
1 213775 1.113439
1 218457 0.393112
or
1 202360 0.315323
1 213775 1.113439
1 218457 0.393112
To control the color of points, add a color column to categorize the data into different groups. Then different colors will be assigned to different groups of data. In this case, column names are compulsory. The name of the first three columns can be any appropriate variable names in R and the order of the first three columns must be fixed as the simplest situation. The name of the color column must be ‘color’.
chr position value color
1 202360 0.315323 a
1 213775 1.113439 a
1 218457 0.393112 a
To control the symbol used for each point, add a shape column. Check http://www.endmemo.com/program/R/pchsymbols.php for more information. In this case, column names are compulsory. The name of the first three columns can be any appropriate variable names in R and the order of the first three columns must be fixed as the simplest situation. The name of the shape column must be ‘shape’.
chr position value shape
1 1 29 15
1 100001 18 15
1 200001 22 15
To control the size of each point, add a size column. Larger number in the size column means lareger point size. In this case, column names are compulsory. The name of the first three columns can be any appropriate variable names in R and the order of the first three columns must be fixed as the simplest situation. The name of the size column must be ‘size’.
chr position value size
1 1 29 1.1
1 100001 18 1.0
1 200001 22 1.1
Users can choose to control two or more of the color, shape and size features at the same time. In this case, column names are compulsory. The name of the first three columns can be any appropriate variable names in R and the order of the first three columns must be fixed as the simplest situation. The name of the shape column must be ‘shape’. The name of the color column must be ‘color’. The name of the size column must be ‘size’. The order of the color, shape and size columns is flexible. Acceptable input data can be
chr position value color shape
1 1 29 a 15
1 100001 18 a 15
1 200001 22 a 15
or
chr position value color shape size
1 1 29 a 15 1.1
1 100001 18 a 15 1.0
1 200001 22 a 15 1.1
or
chr position value color size
1 1 29 a 1.1
1 100001 18 a 1.0
1 200001 22 a 1.1
or
chr position value shape size
1 1 29 15 1.1
1 100001 18 15 1.0
1 200001 22 15 1.1
1.3 Line
The dataset should contain >=3 columns.
In the simplest situation, the dataset should contain 3 columns with fixed order. In this case, column names are optional.
1st column: chromosome ID.
2nd column: chromosome position.
3rd column: data value.
Acceptable input data format can be
chr position value
1 0 0.0428
1 565000 0.0522
1 599000 0.0674
or
1 0 0.0428
1 565000 0.0522
1 599000 0.0674
To add multiple lines and assign different colors to different lines, add a color column to categorize the data into different groups. In this case, column names are compulsory. The name of the first three columns can be any appropriate variable names in R and the order of the first three columns must be fixed as the simplest situation. The name of the color column must be ‘color’.
chr position value color
1 1 29 a
1 100001 18 a
1 1 4 b
1 200001 5 b
1.4 Bar
The dataset should contain >=4 columns.
In the simplest situation, the dataset should contain 4 columns with fixed order. In this case, column names are optional.
1st column: chromosome ID.
2nd column: start coordinate of bars.
3rd column: end coordinate of bars.
4th column: data value.
Acceptable input data format can be
chr start end value
1 1 100000 672
1 100001 200000 486
1 200001 300000 650
or
1 1 100000 672
1 100001 200000 486
1 200001 300000 650
To control the color of bars, add a color column to categorize the data into different groups. Then different colors will be assigned to different groups of data. In this case, column names are compulsory. The name of the first four columns can be any appropriate variable names in R and the order of the first four columns must be fixed as the simplest situation. The name of the color column must be ‘color’.
chr start end value color
1 0 565000 0.5923 a
1 565000 599000 0.6701 a
1 599000 922000 0.6785 a
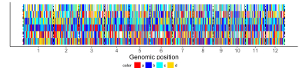
1.5 Rect
The dataset should contain 4 columns with fixed order. Column names are optional.
1st column: chromosome ID.
2nd column: start coordinate of rects.
3rd column: end coordinate of rects.
4th column: data value.
The 4th column can be a character vector or a numeric vector. For a character vector, choose the rect_discrete plot type. For a numeric vector, choose the rect_gradual plot type.
Acceptable input data format can be
chr start end color
1 1 100000 A
1 100001 200000 C
1 200001 300000 A
or
1 1 100000 A
1 100001 200000 C
1 200001 300000 A
or
chr start end NTE
1 1 100000 29
1 100001 200000 18
1 200001 300000 22
or
1 1 100000 29
1 100001 200000 18
1 200001 300000 22
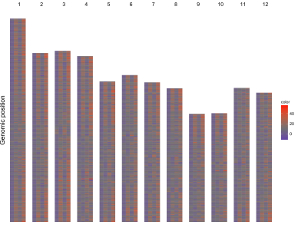
1.6 Heatmap
The dataset should contain >=4 columns. Column names are optional. The order of the first three columns must be fixed as follows.
1st column: chromosome ID.
2nd column: start coordinate of cells.
3rd column: end coordinate of cells.
Except for the first three columns, all the rest columns are treated as data values by shinyChromosome.
The rest columns can be character vectors or numeric vectors. Mix of character vector and numeric vector are not allowed.
For character vectors, choose the heatmap_discrete plot type. For numeric vectors, choose the heatmap_gradual plot type.
Acceptable input data format can be
chr start end val1 val2 val3 val4 val5 val6
1 0 631164 a e c c a b
1 631165 1749192 b b c d d c
1 1749193 2077793 c e a b e e
or
1 0 631164 a e c c a b
1 631165 1749192 b b c d d c
1 1749193 2077793 c e a b e e
or
chr start end TE NTE TR NTR
1 1 100000 4 29 17 45
1 10000001 10100000 9 14 20 28
1 1000001 1100000 1 16 -5 29
or
1 1 100000 4 29 17 45
1 10000001 10100000 9 14 20 28
1 1000001 1100000 1 16 -5 29
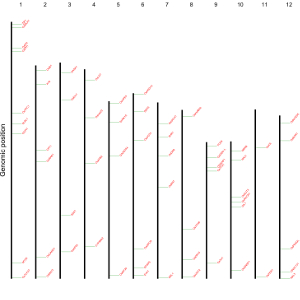
1.7 Segment
The dataset should contain 5 columns with fixed order. Column names are optional.
1st column: chromosome ID.
2nd column: X-axis start coordinate of segments.
3rd column: Y-axis start coordinate of segments.
4th column: X-axis end coordinate of segments.
5th column: Y-axis end coordinate of segments.
Acceptable input data format can be
chr xstart ystart xend yend
1 134291 0 134291 2.8
1 2665412 0 2665412 2.8
1 24392841 0 24392841 2.8
or
1 134291 0 134291 2.8
1 2665412 0 2665412 2.8
1 24392841 0 24392841 2.8
1.8 Text
The dataset should contain 4 columns with fixed order. Column names are optional.
1st column: chromosome ID.
2nd column: X-axis position of texts.
3rd column: Y-axis position of texts.
4th column: the symbols of texts.
Acceptable input data format can be
chr xpos ypos symbol
1 134291 3 OsTLP27
1 2665412 3 MT2D
1 24392841 3 OCPI1
or
1 134291 3 OsTLP27
1 2665412 3 MT2D
1 24392841 3 OCPI1
1.9 Vertical line
The dataset should contain 2 columns with fixed order. Column names are optional.
1st column: chromosome ID.
2nd column: genomic position of vertical lines.
Acceptable input data format can be
chr position
1 0
1 43268879
2 35930381
or
1 0
1 43268879
2 35930381
1.10 Horizontal line
The dataset should contain 1 column. Column names are optional.
1st column: Y-axis value of horizontal lines.
Acceptable input data format can be
position
8
12
5
or
8
12
5
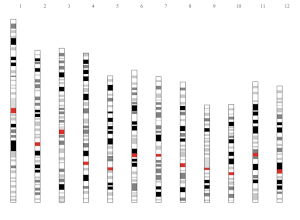
1.11 Ideogram
Ideogram is a schematic representation of chromosomes. Please check https://www.nature.com/scitable/topicpage/chromosome-mapping-idiograms-302 and http://genome.ucsc.edu/cgi-bin/hgTables?db=hg38&hgta_group=map&hgta_track=cytoBand&hgta_table=cytoBand&hgta_doSchema=describe+table+schema for more information. The input data to create ideogram should contain 5 columns with fixed order. Column names are optional.
1st column: chromosome ID.
2nd column: Start coordinate in chromosome sequence.
3rd column: End coordinate in chromosome sequence.
4th column: Name of cytogenetic band.
5th column: Giesma stain results.
Acceptable input data format can be
1 1 399271 p36.33 gneg
1 399271 937418 p36.32 gpos25
1 937418 1249890 p36.31 gneg
2. Two-genome plot
2.1 Data of genome along the horizontal axis
The dataset should contain only 2 columns with fixed order. Column names are optional.
1st column: chromosome ID.
2nd column: chromosome length.
Acceptable input data format can be
chr size
1 43268879
2 35930381
3 36406689
or
1 43268879
2 35930381
3 36406689
2.2 Data of genome along the vertical axis
The dataset should contain only 2 columns with fixed order. Column names are optional.
1st column: chromosome ID.
2nd column: chromosome length.
Acceptable input data format can be
chr size
Chr01 41185095
Chr02 34608401
Chr03 37032663
or
Chr01 41185095
Chr02 34608401
Chr03 37032663
2.3 Point
The dataset should contain >=4 columns.
In the simplest situation, the dataset should contain 4 columns with fixed order. In this case, column names are optional.
1st column: chromosome ID of genome along the horizontal axis.
2nd column: chromosome position in genome along the horizontal axis.
3rd column: chromosome ID of genome along the horizontal axis.
4th column: chromosome position in genome along the vertical axis.
Acceptable input data format can be
chrX posX chrY posY
4 23006000 6 27706220
6 26269000 6 27706227
11 17015000 6 27706228
or
4 23006000 6 27706220
6 26269000 6 27706227
11 17015000 6 27706228
To control the color of points, add a color column. In this case, column names are compulsory. The name of the first four columns can be any appropriate variable names in R and the order of the first four columns must be fixed as the simplest situation. The name of the color column must be ‘color’. The color column can be a character vector or a numeric vector. If the color column is a character vector, choose the point_discrete plot type.
chrX posX chrY posY color
1 15414550 1 17415683 a
1 2314068 1 2291659 a
1 2583523 1 2546654 c
If the color column is a numeric vector, choose the point_gradual plot type.
chrX posX chrY posY color
4 23006000 6 27706220 5.222
6 26269000 6 27706227 10.424
11 17015000 6 27706228 5.802
To control the symbol used for each point, add a shape column. Check http://www.endmemo.com/program/R/pchsymbols.php for more information. In this case, column names are compulsory. The name of the first four columns can be any appropriate variable names in R and the order of the first four columns must be fixed as the simplest situation. The name of the shape column must be ‘shape’. The shape column should be an integer vector.
chrX posX chrY posY shape
1 15414550 1 17415683 12
1 2314068 1 2291659 12
1 2583523 1 2546654 12
To control the size of each point, add a size column. Larger number in the size column means lareger point size. In this case, column names are compulsory. The name of the first four columns can be any appropriate variable names in R and the order of the first four columns must be fixed as the simplest situation. The name of the size column must be ‘size’. The size column should be an integer vector.
chrX posX chrY posY size
1 15414550 1 17415683 1.2
1 2314068 1 2291659 1.2
1 2583523 1 2546654 1.2
Acceptable input data can also be
chrX posX chrY posY color shape
1 15414550 1 17415683 a 12
1 2314068 1 2291659 a 12
1 2583523 1 2546654 c 12
or
chrX posX chrY posY color size
1 15414550 1 17415683 a 1.2
1 2314068 1 2291659 a 1.2
1 2583523 1 2546654 c 1.2
or
chrX posX chrY posY shape size
1 15414550 1 17415683 12 1.2
1 2314068 1 2291659 12 1.2
1 2583523 1 2546654 12 1.2
or
chrX posX chrY posY color shape size
1 15414550 1 17415683 a 12 1.2
1 2314068 1 2291659 a 12 1.2
1 2583523 1 2546654 c 12 1.2
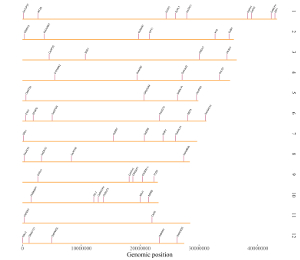
2.4 Segment
The dataset should contain >=6 columns.
In the simplest situation, the dataset should contain 6 columns with fixed order. In this case, column names are optional.
1st column: chromosome ID of genome along the horizontal axis.
2nd column: X-axis start coordinate of segments.
3rd column: X-axis end coordinate of segments.
4th column: chromosome ID of genome along the vertical axis.
5th column: Y-axis start coordinate of segments.
6th column: Y-axis end coordinate of segments.
Acceptable input data can be
chrX startX stopX chrY startY stopY
Chr01 101 21963 Chr01 19600 41490
Chr01 25221 49370 Chr01 41483 65682
Chr01 49604 67964 Chr01 65681 84044
or
Chr01 101 21963 Chr01 19600 41490
Chr01 25221 49370 Chr01 41483 65682
Chr01 49604 67964 Chr01 65681 84044
To control the color of segments, add a color column to categorize data into different groups. Then different colors will be assigned to different groups of data. In this case, column names are compulsory. The name of the first six columns can be any appropriate variable names in R and the order of the first six columns must be fixed as the simplest situation. The name of the color column must be ‘color’.
chrX startX stopX chrY startY stopY color
Chr01 1 35619588 Chr01 1 36185095 a
Chr02 35140161 1 Chr02 34608401 1 b
Chr03 1 33736842 Chr03 37032663 1 c
2.5 Rect
The dataset should contain 7 columns with fixed order. Column names are optional.
1st column: chromosome ID of genome along the horizontal axis.
2nd column: X-axis start coordinate of rects.
3rd column: X-axis end coordinate of rects.
4th column: chromosome ID of genome along the vertical axis.
5th column: Y-axis start coordinate of rects.
6th column: Y-axis end coordinate of rects.
7th column: the color of rects.
The 7th column can be a character vector or a numeric vector. For a character vector, choose the rect_discrete plot type. For a numeric vector, choose the rect_gradual plot type.
Acceptable input data format can be
chrx startx stopx chry starty stopy color
1 1 1000000 1 1 1000000 41
1 1 1000000 1 1000001 2000000 43
1 1 1000000 1 2000001 3000000 59
or
chrx startx stopx chry starty stopy color
1 1 1000000 1 1 1000000 b
1 1 1000000 1 1000001 2000000 b
1 1 1000000 1 2000001 3000000 b
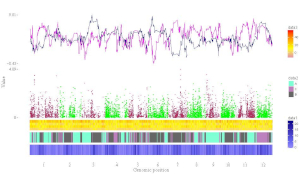
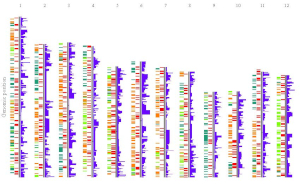
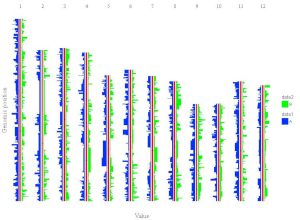
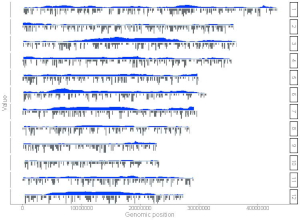
shinyChromosome is a graphical user interface for interactive creation of non-circular whole genome diagrams developed using the R Shiny package.
To create single-genome plot by aligning genome data along all chromosomes of a single genome, go to the Single-genome plot menu.
To cretae two-genome plot for comparison of data across two genomes, go to the Two-genome plot menu.
For the detail format of input data, check the Input data format submenu of the Help menu.
Citation
Yu Y+, Yao W+✉, Wang Y, Huang F. shinyChromosome: An R/Shiny Application for Interactive Creation of Non-circular Plots of Whole Genomes. Genomics, Proteomics & Bioinformatics, 2020 (+ co-first author)
Software references
- R Development Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna. R version 3.5.0 (2018)
- RStudio and Inc. shiny: Web Application Framework for R. R package version 1.0.5 (2017)
- Lionel Henry and Hadley Wickham. rlang: Functions for Base Types and Core R and “Tidyverse” Features. R package version 0.2.1 (2018)
- H. Wickham. ggplot2: Create Elegant Data Visualisations Using the Grammar of Graphics. R package version 3.0.0 (2018)
- Gábor Csárdi, Kuba Podgórski, Rich Geldreich. zip: Cross-Platform zip Compression. R package version 2.0.2 (2019)
- Erich Neuwirth. RColorBrewer: ColorBrewer palettes. R package version 1.1-2 (2014)
- Hadley Wickham. plyr: Tools for Splitting, Applying and Combining Data. R package version 1.8.4 (2016)
- Jeffrey B. Arnold. ggthemes: Extra Themes, Scales and Geoms for “ggplot2”. R package version 3.4.0 (2017)
- Christoph Burow, Urs Tilmann Wolpert and Sebastian Kreutzer. RLumShiny: “Shiny” Applications for the R Package “Luminescence”. R package version 0.2.0 (2017)
- Baptiste Auguie. gridExtra: Miscellaneous Functions for “Grid” Graphics. R package version 2.3 (2017)
- Hadley Wickham. reshape2: Flexibly Reshape Data: A Reboot of the Reshape Package. R package version 1.4.3 (2017)
- Matt Dowle and Arun Srinivasan. data.table: Extension of “data.frame”. R package version 1.10.4-3 (2017)
- JJ Allaire, Jeffrey Horner, Vicent Marti and Natacha Porte. markdown: “Markdown” Rendering for R. R package version 0.8 (2017)
Further references
This application was created by Yiming Yu and Wen Yao. Please send bugs and feature requests to Yiming Yu (yimingyyu at gmail.com) or Wen Yao (venyao at qq.com). This application uses the shiny package from RStudio.
Note
For Mac users, we recommend using shinyChromosome with the Google Chrome browser or other browsers developed based on Chrominum.